Research
Research Thrust: Physics-informed Knowledge Fusion in Genomic Foundation Models
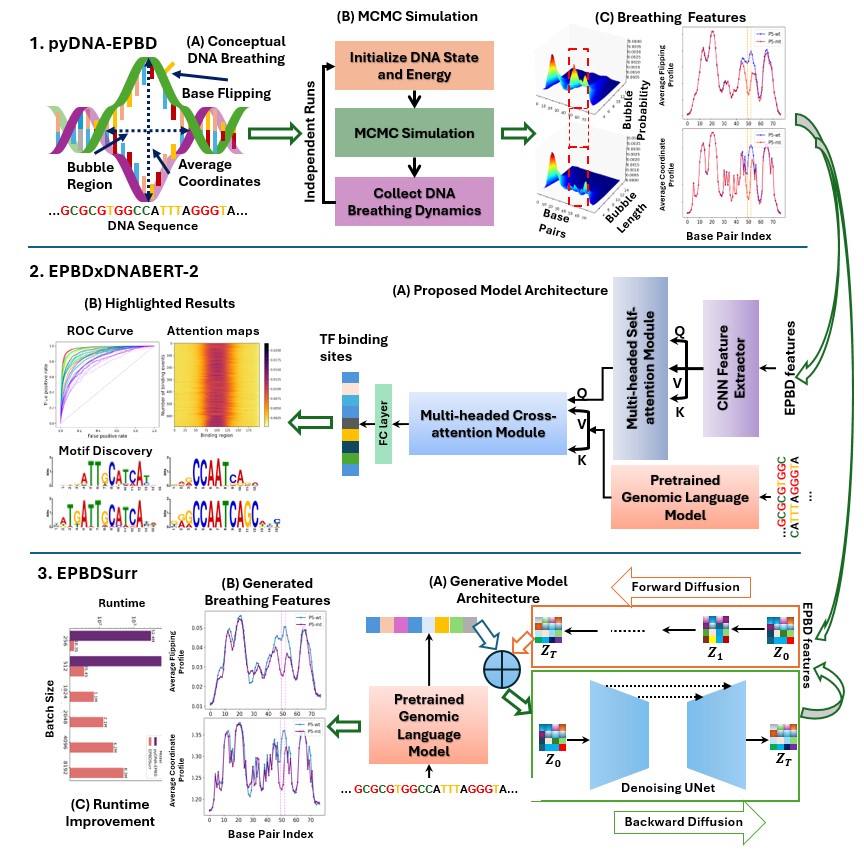
DNA double helical structure unwinds and recombines locally due to the DNA thermal fluctuations. We develop a physics-guided simulation tool to model and capture this phenomena. Next, we design a deep learning based multi-modal architecture that integrates a genomic foundation model and DNA breathing features extracted by the simulation for predicting transcription factor binding sites across various cell lines. To expediate the generation of DNA breathing features, we develop a conditional generative architecture that is highly efficient and accurate with respect to the simulation.
Publications:
- Kabir, A., Bhattarai, M., Rasmussen, K. Ø., Shehu, A., Usheva, A., Bishop, A. R., & Alexandrov, B. (2023). Examining DNA breathing with pyDNA-EPBD. Bioinformatics (Oxford, England), 39(11). doi:10.1093/bioinformatics/btad699
- Kabir, A., Bhattarai, M., Peterson, S., Najman-Licht, Y., Rasmussen, K. Ø., Shehu, A., … Usheva, A. (2024). DNA breathing integration with deep learning foundational model advances genome-wide binding prediction of human transcription factors. Nucleic Acids Research, 52(19), e91. doi:10.1093/nar/gkae783
- Kabir, A., Inan, T. T., Rasmussen, K., Shehu, A., Usheva, A., Bishop, A., … Bhattarai, M. (2024). Scalable DNA feature generation and transcription factor binding prediction via deep surrogate models. doi:10.1101/2024.12.06.626709
- Inan, T. T., Kabir, A., Rasmussen, K., Shehu, A., Usheva, A., Bishop, A., Alexandrov, B., & Bhattarai, M. (2024). Efficient High-Throughput DNA Breathing Features Generation Using Jax-EPBD. doi:10.1101/2024.12.06.627191
Research Thrust: Biological Sequence is Necessary but not Sufficient for Accurate Predictions
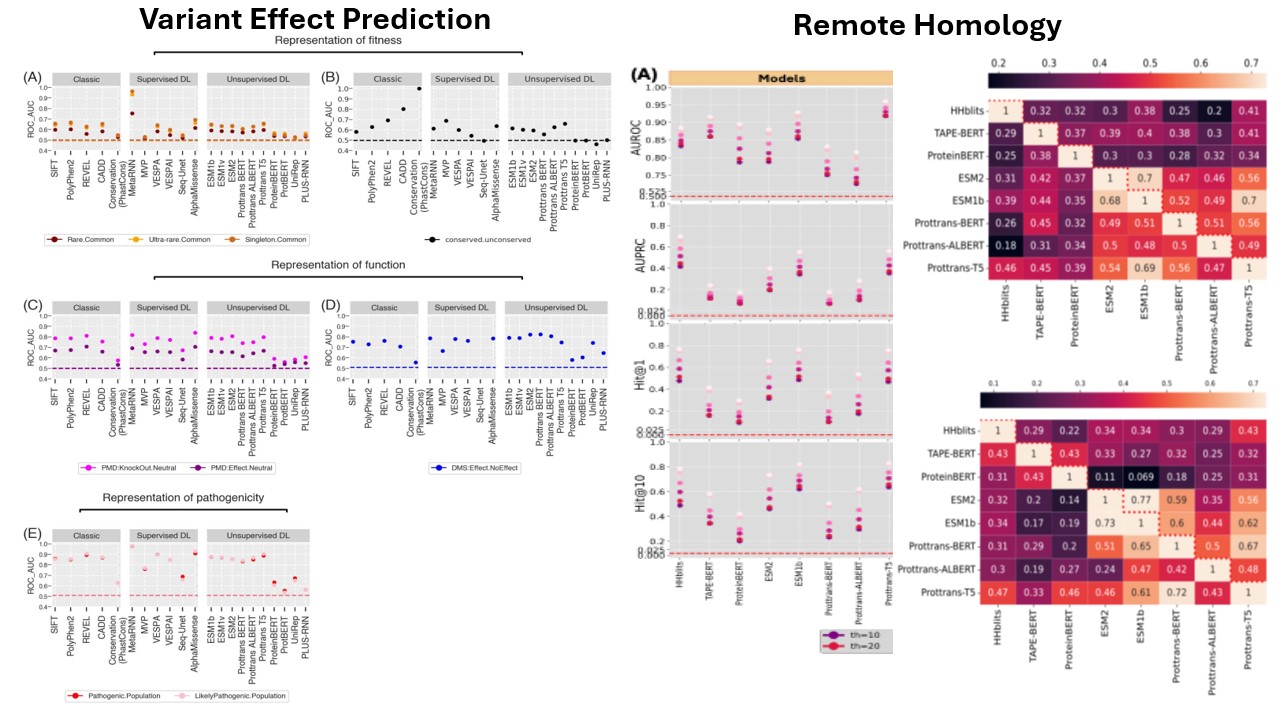
A class of foundation models have been proposed by addressing many suitable biologically relevant prediction problems. We assess a set of state of the art methods in terms of functional characterization of the molecules. We highlight our findings in mutation effect and remote homology understanding of the methods as a stress test of capturing the tiny changes in the molecules.
Publications:
- Kabir, A., Moldwin, A., & Shehu, A. (2023). A comparative analysis of transformer-based protein language models for remote homology prediction. Proceedings of the 14th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, 1–9. Presented at the BCB ’23: 14th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, Houston TX USA. doi:10.1145/3584371.3612942
- Kabir, A., Moldwin, A., Bromberg, Y., & Shehu, A. (2024). In the twilight zone of protein sequence homology: do protein language models learn protein structure? Bioinformatics Advances, 4(1), vbae119. doi:10.1093/bioadv/vbae119
- Bromberg, Y., Prabakaran, R., Kabir, A., & Shehu, A. (2024). Variant effect prediction in the age of machine learning. Cold Spring Harbor Perspectives in Biology, 16(7), a041467. doi:10.1101/cshperspect.a041467
Research Thrust: Integration of Biological Multi-modalities
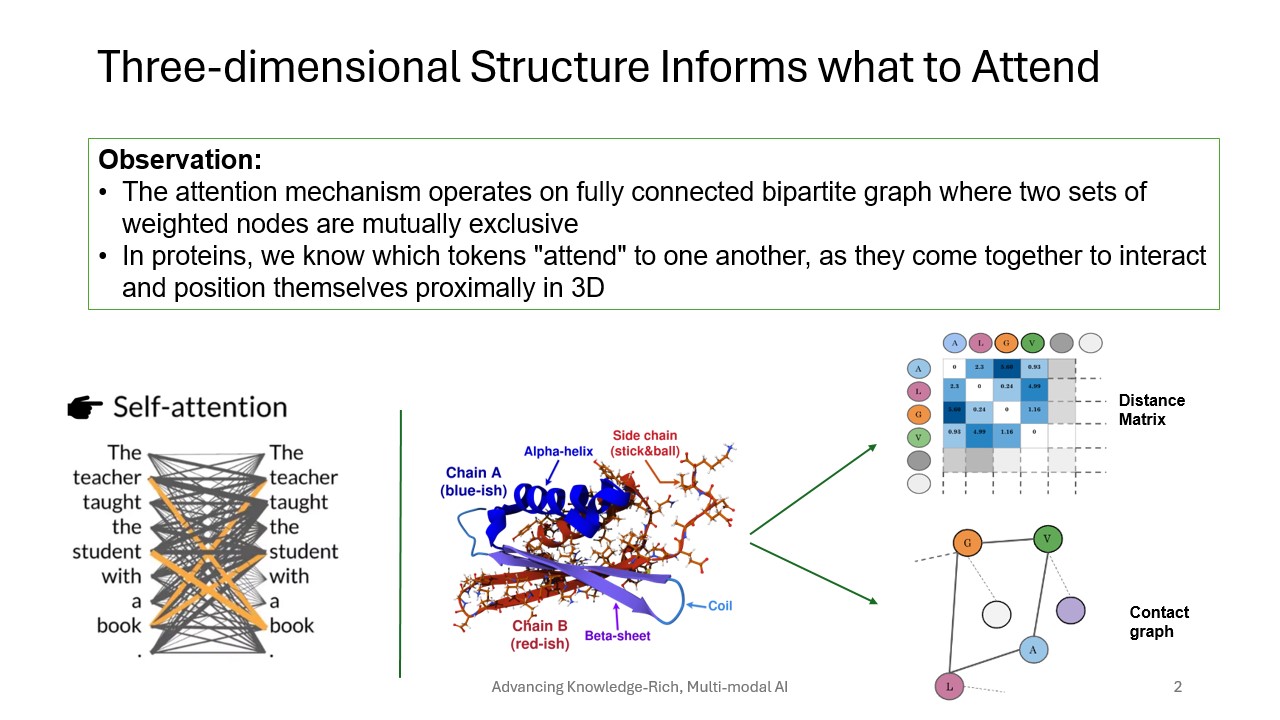
We design and develop effective deep learning based architectures by incorporating multi-modal representations of the molecules for predicting biological tasks, such as protein function prediction, structure prediction.
Publications:
- Kabir, A., & Shehu, A. (2022). GOProFormer: A multi-modal transformer method for Gene Ontology protein function prediction. Biomolecules, 12(11), 1709. doi:10.3390/biom12111709
- Kabir, A., & Shehu, A. (2022, November). Sequence-structure embeddings via protein language models improve on prediction tasks. 2022 IEEE International Conference on Knowledge Graph (ICKG). Presented at the 2022 IEEE International Conference on Knowledge Graph (ICKG), Orlando, FL, USA. doi:10.1109/ickg55886.2022.00021
- Du, Y., Kabir, A., Zhao, L., & Shehu, A. (2020). From interatomic distances to protein tertiary structures with a deep convolutional neural network. Proceedings of the 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics. Presented at the BCB ’20: 11th ACM International Conference on Bioinformatics, Computational Biology and Health Informatics, Virtual Event USA. doi:10.1145/3388440.3414699